3T3 Mouse Fibroblast Proteomics is a free, stand-alone Windows program. It is provided “as-is”, without any express or implied warranty.
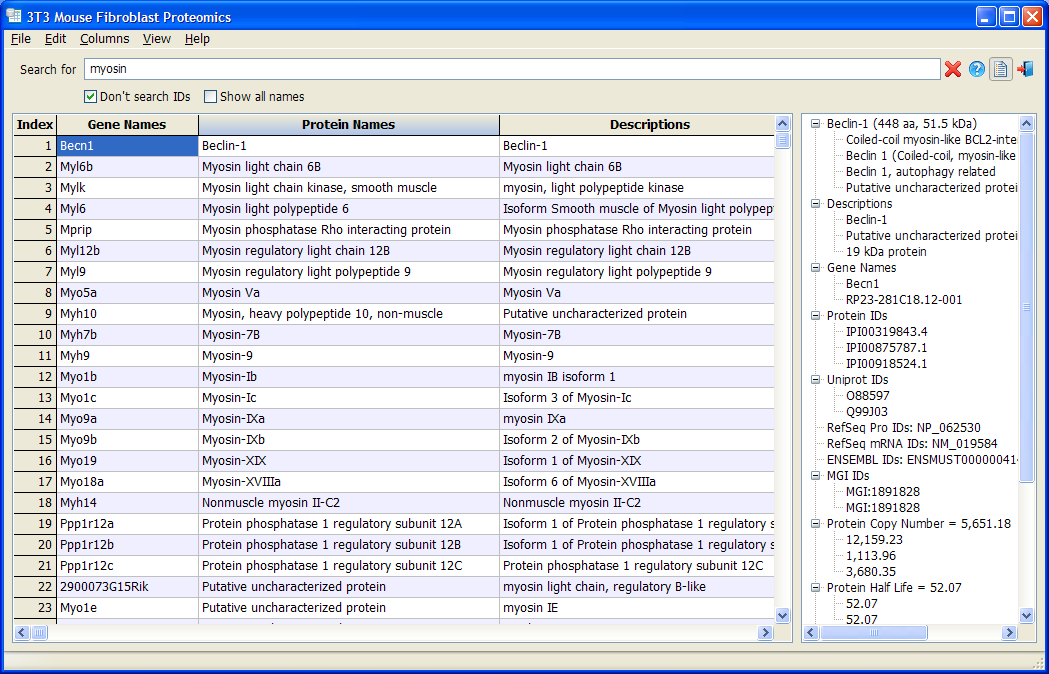
Proteomics3T3 is a database based on Schwanhäusser et. al., Nature 2011 and its Corrigendum. The original database is in an Excel file and is fairly difficult to read, search, sort or compare. Proteomics3T3 puts all data in a customizable table. Users are allowed to hide or show each column, change the font and colors, and sort the data with any column. A data panel shows all the detail information of a gene in an organization tree view. The data panel can be hidden or can be undocked from the main window.
Proteomics3T3 provides powerful filtering of the data. Simply type in the text box and only genes containing the text will be shown. Different filters can be combined using Boolean searching. For examples, “Myosin | Dynein | Kinesin” shows all molecular motors; “GTPase /rho” shows all non-Rho GTPases.
Proteomics3T3 is a standalone program with the database embedded in the executable. It does not require installation and can be safely removed by deleting the file.
Reference
-
Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M.
Global quantification of mammalian gene expression control.
Nature,
2011;
473(7347):337-42.


